Lesion mimic mutants (LMMs) constitutively and spontaneously display genetic lesions (necrotic spots of dead cells) in the absence of pathogenic infection, identical to disease-caused lesions due to likeness between the non-pathogenic and pathogenic syndromes associated with hypersensitive response and cell death. The broad-spectrum, nonrace-specific resistance expressed by these mutants to fungal and bacterial rice pathogens have attracted the attention to use the genetic mechanism for developing disease resistant plants (Zeng et al. 2004). However, direct use of the mutated genes in breeding for disease resistance have not met with success, as they result in abnormal phenotypes (Yin et al. 2000). In a land mark study (Sha et al. 2023), an international team of researchers jointly led by Pamela C. Ronald from the University of California, Davis, USA and Guotian Li from Huazhong Agricultural University in Wuhan, China has isolated an LMM from a fast neutron (FN)-induced mutant population of a japonica rice cultivar KitaakeX (a derivative of Kitaake) carrying a bacterial blight resistance gene Xa21 named rbl1 that has resulted due to a 29-base-pair deletion in the gene Resistance to Blast 1 (RBL1). The rbl1 mutant exhibits broad-spectrum disease resistance to both blast (Magnaporthe oryzae) and bacterial blight (Xanthomonas oryzae pv. oryzae) due to pathogen-triggered accumulation of reactive oxygen species (ROS) , elevated levels of salicylic acid and upregulation of plant defense-related genes coupled with drastic reduction in yield. The highly conserved disease resistance genes, Lr34 (Boni et al. 2018) and mlo (mildew locus O, Gruner et al. 2020) used in different crops for disease resistance are also from LMMs. RBL is expressed in all the tissues of rice plant with higher intensity in the leaves, and it is inducible by the blast fungus M. oryzae. WT RBL1 encodes an enzyme, cytidine diphosphate diacylglycerol (CDP-DAG) synthase (CDS) required for phospholipid biosynthesis leading to the accumulation of an established susceptibility factor phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) in rice (Fig. 1). PtdIns(4,5)P2 is a minor phospholipid component of cell membranes. This susceptibility factor gets enriched in the membrane covering the cellular structures of invasive hyphae and biotrophic interfacial complex (BIC) as found in the extra-invasive hyphal membrane (EIHM) in the hemibiotrophic Colletotrichum-Arabidopsis (Shimada et al. 2019) and in the extra-haustorial membrane (EHM) in the biotrophic Erysiphe-Arabidopsis (Qin et al. 2020) pathosystems favouring disease susceptibility. Both EIHM and EHM are continuous extension of the plant plasma membrane. PtdIns(4,5)P2 is associated with effector secretion and fungal infection to promote disease susceptibility.
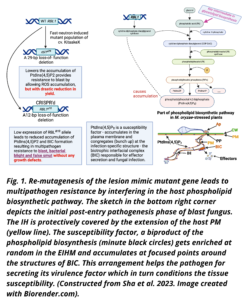
Disrupting the function of the susceptibility factor by mutating the cognate gene leads to disease resistance, usually with more durability and broad-spectrum effect preventing infection by a wide range of pathogen strains as well by more than one pathogen species.
The 2020 Nobel prize (Chemistry) winning genome editing technology (Doudna and Charpentier) discovered in the year 2012 arose from the adaptive immune system of prokaryotes (bacteria and archaea) which protect them from invasive viruses and plasmids by silencing their foreign nucleic acids (DNA). CRISPR (clustered regularly interspaced short palindromic repeats) RNAs (termed crRNAs) recruit a Cas9 (CRISPR-associated 9) endonuclease for this function. CRISPR-Cas9 genome editing technology has profoundly influenced biological research especially with genetically inherited human diseases and agricultural practices and products. CRISPR-Cas9 technique has become the commonly used technology facilitating insertion, deletion or modifying the genes in the plant genome to achieve desired traits. This also greatly helps in understanding the gene functions and their validations for experimental purposes.
Sha et al. (2023) by targeting the wild type (WT) RBL1 gene with genome editing CRISPR technology obtained an allele of RBL1 named RBL1Δ12 caused by a 12-bp deletion. This has resulted in a sort of metabolic engineering

fine-tuning the phospholipid metabolism of the plants for reducing the level of the susceptibility factor, PtdIns(4,5)P2 by interfering in the phospholipid biosynthetic pathway of the plants.
Lowering this susceptibility factor confers multipathogen, broad-spectrum resistance to important hemibiotrophic rice pathogens causing blast, bacterial blight (including for strains virulent on Xa21), and false smut (Ustilaginoidea virens) together (Fig.2), without causing any yield penalty in the model rice variety, Kitaake under experimental and in small-scale yield trials. The consistency of disease resistance combined with normal phenotypes of RBL1 CRISPR-edited lines has been demonstrated with two other rice cultivars, Nipponbare and Zhonghua 11. The multi-pathogen resistance effective against more than one pathogen species is highly preferable in agricultural production. The RBL1Δ12 allele with multipathogen resistance trait including for a difficult-to-manage flower-infecting pathogen for which resistance genes have not been located so far in rice germplasm, together with high yield is a valuable genetic resource for rice improvement in the plant breeder’s arsenal.
References
Boni R, Chauhan H, Hensel G, Roulin A, Sucher J, Kumlehn J, Brunner S, Krattinger SG, Keller B. Pathogen-inducible Ta-Lr34res expression in heterologous barley confers disease resistance without negative pleiotropic effects. Plant Biotechnol. J. 16,245-253.
Gruner, K, Esser T, Acevedo-Garcia J, Freh M, Habig M, Strugala R, Stukenbrock E, Ulrich Schaffrath U,
Panstruga R. 2020. Evidence for allele-specific levels of enhanced susceptibility of wheat mlo mutants to the
hemibiotrophic fungal pathogen Magnaporthe oryzae pv. Triticum. Genes 11, 517.
Qin L, Zhou Z, Li Q, Zhai C, Liu L, Quilichini TD, Gao P, Kessler SA, Jaillais Y, Datla R, Peng G, Xiang D,
Wei Y. 2020. Specific recruitment of phosphoinositide species to the plant-pathogen interfacial membrane
underlies Arabidopsis susceptibility to fungal infection. Plant Cell 32,1665–1688.
Sha G, Sun P, Kong X, Han X, Sun Q, Fouillen L, Zhao J, Li Y, Yang L, Wang Y, Gong Q, Zhou Y, Zhou
W, Jain R, Gao J, Huang R, Chen X, Zheng L, Zhang W, Qin Z, Zhou Q, Zeng O, Xie K, Xu J, Chiu
T-Y, Guo L, Mortimer JC, Boutté Y, Li Q, Kang Z, Ronald PC, Li G. 2023. Genome editing of a
rice CDP-DAG synthase confers multipathogen resistance. Nature 618,1017-1023.
Shimada T, Betsuyaku S, Inada N, Ebine K, Fujimoto M, Uemura T, Takano Y, Fukuda4 H, Nakano A,
Ueda T. 2019. Enrichment of phosphatidylinositol 4,5-bisphosphate in the extra-invasive hyphal
membrane promotes Colletotrichum infection of Arabidopsis thaliana. Plant Cell Physiol. 60,1514–1524.
Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush GS, Wang G-L. 2000. Characterizing rice lesion mimic
mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol.
Plant-Microbe Intrctn. 1,869-876.
Zeng L-R, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang G-L.
Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat
protein endowed with E3 ubiquitin ligase activity. Plant Cell 16,2795–2808
Suggested citation: Sridhar, R. (2023, August 25) First CRISPR’d rice with multi-pathogen resistance.
https://sridharr.com/first-crisprd-rice-with-multi-pathogen-resistance/
DOI: 10.13140/RG.2.2.30469.12001.